Yaffe, Theo Cleland; James G. Mainprize; Olivier Alonzo-Proulx; Jennifer A. Harvey; Roberta A Jong; Anne L. Martel; Martin J.: Use of convolutional neural networks to predict risk of masking by mammographic density. SPIE Medical Imaging, 2019, Computer-Aided Diagnosis, vol. 10950, 2019. @conference{Yaffe2019,
title = {Use of convolutional neural networks to predict risk of masking by mammographic density},
author = {Theo Cleland; James G. Mainprize; Olivier Alonzo-Proulx; Jennifer A. Harvey; Roberta A Jong; Anne L. Martel; Martin J. Yaffe},
year = {2019},
date = {2019-03-13},
booktitle = { SPIE Medical Imaging, 2019, Computer-Aided Diagnosis},
volume = {10950},
abstract = {Sensitivity of screening mammography is reduced by increased mammographic density (MD). MD can obscure or “mask” developing lesions making them harder to detect. Predicting masking risk may be an effective tool for a stratified screening program where selected women can receive alternative screening modalities that are less susceptible to masking. Here, we investigate whether the use of artificial intelligence can accurately predict the masking risk and compare its performance to that of conventional BI-RADS density classification. The analysis was based on mammograms of 214 subjects comprised of 147 women with a screen-detected (SD) or “non-masked” cancer and 67 that developed a non-screen detected (NSD) or presumably masked cancer within 2 years following a negative screen. Prior to analysis, mammograms were pre-processed into quantitative MD maps using an in-house algorithm. A transfer learning approach was used to train a convolutional neural network (CNN) based on VGG-16 in a seven cross-fold approach to classify masking status. A two-step transfer learning method was also used where the pre-trained CNN was initially trained on 5,865 mammograms to classify by BI-RADS density category and then trained for masking status. Using BI-RADS density as a masking risk predictor has an AUC of 0.64 [0.57 - 0.71 95CI]. The CNN-mask yielded an AUC of 0.76 [0.68 - 0.81]. Combining the CNN-mask with our previous hand-crafted masking risk predictor, the AUC improved to 0.78 [0.70 - 0.83]. The combined AUC improved to 0.81 [0.72-0.90] when analysis was restricted to NSD cancers surfacing clinically within one year after a negative screen. The two-step transfer learning yielded similar performance. This work suggests that a CNN masking risk predictor can be used to guide a stratified screening program to overcome the limitations of screening mammography in dense breasts.},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}
Sensitivity of screening mammography is reduced by increased mammographic density (MD). MD can obscure or “mask” developing lesions making them harder to detect. Predicting masking risk may be an effective tool for a stratified screening program where selected women can receive alternative screening modalities that are less susceptible to masking. Here, we investigate whether the use of artificial intelligence can accurately predict the masking risk and compare its performance to that of conventional BI-RADS density classification. The analysis was based on mammograms of 214 subjects comprised of 147 women with a screen-detected (SD) or “non-masked” cancer and 67 that developed a non-screen detected (NSD) or presumably masked cancer within 2 years following a negative screen. Prior to analysis, mammograms were pre-processed into quantitative MD maps using an in-house algorithm. A transfer learning approach was used to train a convolutional neural network (CNN) based on VGG-16 in a seven cross-fold approach to classify masking status. A two-step transfer learning method was also used where the pre-trained CNN was initially trained on 5,865 mammograms to classify by BI-RADS density category and then trained for masking status. Using BI-RADS density as a masking risk predictor has an AUC of 0.64 [0.57 - 0.71 95CI]. The CNN-mask yielded an AUC of 0.76 [0.68 - 0.81]. Combining the CNN-mask with our previous hand-crafted masking risk predictor, the AUC improved to 0.78 [0.70 - 0.83]. The combined AUC improved to 0.81 [0.72-0.90] when analysis was restricted to NSD cancers surfacing clinically within one year after a negative screen. The two-step transfer learning yielded similar performance. This work suggests that a CNN masking risk predictor can be used to guide a stratified screening program to overcome the limitations of screening mammography in dense breasts. |
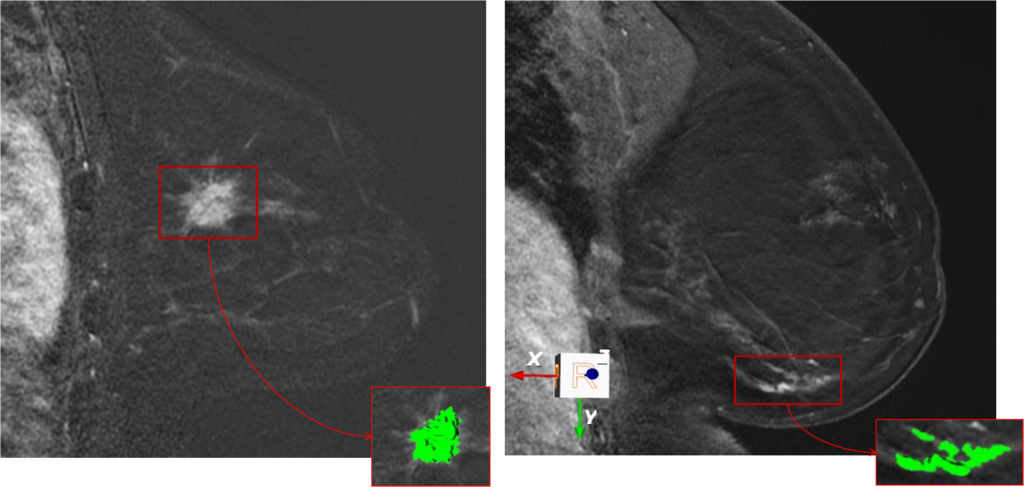
Kuling, Grey; Fashandi, Homa; Lu, YingLi; Wu, Hongbo; Martel, Anne L.: Breast Volume and Fibroglandular Tissue Segmentation in MRI using a Deep Learning Unet. ISMRM Workshop on Breast MRI: Advancing the State of the Art, 2018. @workshop{Kuling2018,
title = {Breast Volume and Fibroglandular Tissue Segmentation in MRI using a Deep Learning Unet},
author = {Grey Kuling and Homa Fashandi and YingLi Lu and Hongbo Wu and Anne L. Martel},
url = {http://martellab.com/wp-content/uploads/2019/09/GCK_ISMRMAbstract_DLSegmatation_072018-3.pdf},
year = {2018},
date = {2018-09-10},
urldate = {2018-09-10},
booktitle = {ISMRM Workshop on Breast MRI: Advancing the State of the Art},
keywords = {},
pubstate = {published},
tppubtype = {workshop}
}
|
Wu, Hongbo: Automatic Computer Aided Diagnosis of Breast Cancer in Dynamic Contrast Enhanced Magnetic Resonance Images. University of Toronto, Department Medical Biophysics , 2016. @mastersthesis{Wu2016,
title = {Automatic Computer Aided Diagnosis of Breast Cancer in Dynamic Contrast Enhanced Magnetic Resonance Images},
author = {Wu, Hongbo},
url = {http://hdl.handle.net/1807/76226},
year = {2016},
date = {2016-11-01},
address = {Department Medical Biophysics },
school = {University of Toronto},
abstract = {Automated Computer Aided Diagnosis (CADx) systems have the potential to improve the diagnostic accuracy of radiologists. Most CADx algorithms use features generated from outlined regions to differentiate between benign and malignant lesions. Manually outlining these regions for the purpose of analysis is not viable and therefore an automated segmentation method is essential. Our proposed method uses a trained deep Artificial Neural Network (ANN) to classify overlapping tiles in breast Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) images as lesion or non-lesion. The classified tiles are then grouped into regions. Additional morphological, kinetic and textural features are computed for each detected region. A cascaded Random Forests Classifier (RFC) classifies the regions as malignant or benign. Our method was tested on a dataset containing 71 malignant, 140 benign, and 316 normal studies. Free-response Receiver Operating Characteristic (FROC) analysis of our method shows 94.4% sensitivity at 0.12 false positive detections per normal study.},
keywords = {},
pubstate = {published},
tppubtype = {mastersthesis}
}
Automated Computer Aided Diagnosis (CADx) systems have the potential to improve the diagnostic accuracy of radiologists. Most CADx algorithms use features generated from outlined regions to differentiate between benign and malignant lesions. Manually outlining these regions for the purpose of analysis is not viable and therefore an automated segmentation method is essential. Our proposed method uses a trained deep Artificial Neural Network (ANN) to classify overlapping tiles in breast Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) images as lesion or non-lesion. The classified tiles are then grouped into regions. Additional morphological, kinetic and textural features are computed for each detected region. A cascaded Random Forests Classifier (RFC) classifies the regions as malignant or benign. Our method was tested on a dataset containing 71 malignant, 140 benign, and 316 normal studies. Free-response Receiver Operating Characteristic (FROC) analysis of our method shows 94.4% sensitivity at 0.12 false positive detections per normal study. |
Gallego, Cristina: Automatic 3D Segmentation of the Breast in MRI. University of Toronto, Medical Biophysics, 2011. @mastersthesis{Gallego2011,
title = {Automatic 3D Segmentation of the Breast in MRI},
author = {Gallego, Cristina},
url = {http://hdl.handle.net/1807/30619},
year = {2011},
date = {2011-12-08},
address = {Medical Biophysics},
school = {University of Toronto},
abstract = {Breast cancer is currently the most common diagnosed cancer among women and a significant cause of death. Breast density is considered a significant risk factor and an important biomarker influencing the later risk of breast cancer. Therefore, ongoing epidemiological studies using MRI are evaluating quantitatively breast density in young women. One of the challenges is segmenting the breast in order to calculate total breast volume and exclude non-breast surrounding tissues. This thesis describes an automatic 3D breast volume segmentation based on 3D local edge detection using phase congruency and Poisson surface reconstruction to extract the total breast volume in 3D. The boundary localization framework is integrated on a subsequent atlas-based segmentation using a Laplacian framework. The 3D segmentation achieves breast-air and breast-chest wall boundary localization errors with a median of 1.36 mm and 2.68 mm respectively when tested on 409 MRI datasets.
},
keywords = {},
pubstate = {published},
tppubtype = {mastersthesis}
}
Breast cancer is currently the most common diagnosed cancer among women and a significant cause of death. Breast density is considered a significant risk factor and an important biomarker influencing the later risk of breast cancer. Therefore, ongoing epidemiological studies using MRI are evaluating quantitatively breast density in young women. One of the challenges is segmenting the breast in order to calculate total breast volume and exclude non-breast surrounding tissues. This thesis describes an automatic 3D breast volume segmentation based on 3D local edge detection using phase congruency and Poisson surface reconstruction to extract the total breast volume in 3D. The boundary localization framework is integrated on a subsequent atlas-based segmentation using a Laplacian framework. The 3D segmentation achieves breast-air and breast-chest wall boundary localization errors with a median of 1.36 mm and 2.68 mm respectively when tested on 409 MRI datasets.
|
Levman, Jacob: Pattern Recognition Applied to the Computer-aided Detection and Diagnosis of Breast Cancer from Dynamic Contrast-enhanced Magnetic Resonance Breast Images. University of Toronto, 2010. @phdthesis{Levman2010,
title = {Pattern Recognition Applied to the Computer-aided Detection and Diagnosis of Breast Cancer from Dynamic Contrast-enhanced Magnetic Resonance Breast Images},
author = {Jacob Levman },
url = {http://hdl.handle.net/1807/24361},
year = {2010},
date = {2010-04-21},
urldate = {2010-04-21},
address = {Medical Biophysics},
school = {University of Toronto},
abstract = {The goal of this research is to improve the breast cancer screening process based on magnetic resonance imaging (MRI). In a typical MRI breast examination, a radiologist is responsible for visually examining the MR images acquired during the examination and identifying suspect tissues for biopsy. It is known that if multiple radiologists independently analyze the same examinations and we biopsy any lesion that any of our radiologists flagged as suspicious then the overall screening process becomes more sensitive but less specific. Unfortunately cost factors prohibit the use of multiple radiologists for the screening of every breast MR examination. It is thought that instead of having a second expert human radiologist to examine each set of images, that the act of second reading of the examination can be performed by a computer-aided detection and diagnosis system. The research presented in this thesis is focused on the development of a computer-aided detection and diagnosis system for breast cancer screening from dynamic contrast-enhanced magnetic resonance imaging examinations. This thesis presents new computational techniques in supervised learning, unsupervised learning and classifier visualization. The techniques have been applied to breast MR lesion data and have been shown to outperform existing methods yielding a computer aided detection and diagnosis system with a sensitivity of 89% and a specificity of 70%.},
keywords = {},
pubstate = {published},
tppubtype = {phdthesis}
}
The goal of this research is to improve the breast cancer screening process based on magnetic resonance imaging (MRI). In a typical MRI breast examination, a radiologist is responsible for visually examining the MR images acquired during the examination and identifying suspect tissues for biopsy. It is known that if multiple radiologists independently analyze the same examinations and we biopsy any lesion that any of our radiologists flagged as suspicious then the overall screening process becomes more sensitive but less specific. Unfortunately cost factors prohibit the use of multiple radiologists for the screening of every breast MR examination. It is thought that instead of having a second expert human radiologist to examine each set of images, that the act of second reading of the examination can be performed by a computer-aided detection and diagnosis system. The research presented in this thesis is focused on the development of a computer-aided detection and diagnosis system for breast cancer screening from dynamic contrast-enhanced magnetic resonance imaging examinations. This thesis presents new computational techniques in supervised learning, unsupervised learning and classifier visualization. The techniques have been applied to breast MR lesion data and have been shown to outperform existing methods yielding a computer aided detection and diagnosis system with a sensitivity of 89% and a specificity of 70%. |
Levman, Jacob; Leung, Tony; Causer, Petrina; Plewes, Don; Martel, Anne L.: Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. In: IEEE Transactions on Medical Imaging , vol. 27, no. 5, pp. 688 - 696, 2008. @article{Levman2008,
title = {Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines},
author = {Jacob Levman and Tony Leung and Petrina Causer and Don Plewes and Anne L. Martel},
url = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2891012/},
doi = {10.1109/TMI.2008.916959},
year = {2008},
date = {2008-05-01},
journal = {IEEE Transactions on Medical Imaging },
volume = {27},
number = {5},
pages = {688 - 696},
abstract = {Early detection of breast cancer is one of the most important factors in determining prognosis for women with malignant tumors. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has been shown to be the most sensitive modality for screening high-risk women. Computer-aided diagnosis (CAD) systems have the potential to assist radiologists in the early detection of cancer. A key component of the development of such a CAD system will be the selection of an appropriate classification function responsible for separating malignant and benign lesions. The purpose of this study is to evaluate the effects of variations in temporal feature vectors and kernel functions on the separation of malignant and benign DCE-MRI breast lesions by support vector machines (SVMs). We also propose and demonstrate a classifier visualization and evaluation technique. We show that SVMs provide an effective and flexible framework from which to base CAD techniques for breast MRI, and that the proposed classifier visualization technique has potential as a mechanism for the evaluation of classification solutions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Early detection of breast cancer is one of the most important factors in determining prognosis for women with malignant tumors. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has been shown to be the most sensitive modality for screening high-risk women. Computer-aided diagnosis (CAD) systems have the potential to assist radiologists in the early detection of cancer. A key component of the development of such a CAD system will be the selection of an appropriate classification function responsible for separating malignant and benign lesions. The purpose of this study is to evaluate the effects of variations in temporal feature vectors and kernel functions on the separation of malignant and benign DCE-MRI breast lesions by support vector machines (SVMs). We also propose and demonstrate a classifier visualization and evaluation technique. We show that SVMs provide an effective and flexible framework from which to base CAD techniques for breast MRI, and that the proposed classifier visualization technique has potential as a mechanism for the evaluation of classification solutions. |