Kuling, Grey; Belinda, Curpen; Martel, Anne L.: Accurate estimation of density and background parenchymal enhancement in breast MRI using deep regression and transformers. In: Li, Hui; Giger, Maryellen L.; Drukker, Karen; Whitney, Heather M. (Ed.): pp. 22, SPIE, 2024, ISBN: 9781510680203. @inproceedings{Kuling2024,
title = {Accurate estimation of density and background parenchymal enhancement in breast MRI using deep regression and transformers},
author = {Grey Kuling and Curpen Belinda and Anne L. Martel},
editor = {Hui Li and Maryellen L. Giger and Karen Drukker and Heather M. Whitney},
doi = {10.1117/12.3025341},
isbn = {9781510680203},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {17th International Workshop on Breast Imaging (IWBI 2024)},
pages = {22},
publisher = {SPIE},
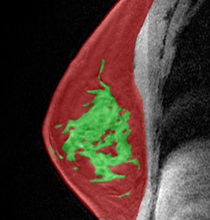
abstract = {Early detection of breast cancer is important for improving survival rates. Based on accurate and tissue-specific risk factors, such as breast density and background parenchymal enhancement (BPE), risk-stratified screening can help identify high-risk women and provide personalized screening plans, ultimately leading to better outcomes. Measurements of density and BPE are carried out through image segmentation, but volumetric measurements may not capture the qualitative scale of these tissue-specific risk factors. This study aimed to create deep regression models that estimate the interval scale underlying the BI-RADS density and BPE categories. These models incorporate a 3D convolutional encoder and transformer layers to comprehend time-sequential data in DCE-MRI. The correlation between the models and the BI-RADS categories was evaluated with Spearman coefficients. Using 1024 patients with a BI-RADS assessment score of 3 or less and no prior history of breast cancer, the models were trained on 50% of the data and tested on 50%. The density and BPE ground truth labels were extracted from the radiology reports using BI-RADS BERT. The ordinal classes were then translated to a continuous interval scale using a linear link function. The density regression model is strongly correlated to the BI-RADS category with a correlation of 0.77, slightly lower than segmentation %FGT. The BPE regression model with transformer layers shows a moderate correlation with radiologists at 0.52, similar to the segmentation %BPE. The deep regression transformer has an advantage over segmentation as it doesn’t need time-point image registration, making it easier to use.
(2024) Published by SPIE. Downloading of the abstract is permitted for personal use only.
Citation Download Citation
Grey Kuling, Belinda Curpen, and Anne L . Martel "Accurate estimation of density and background parenchymal enhancement in breast MRI using deep regression and transformers"},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
Early detection of breast cancer is important for improving survival rates. Based on accurate and tissue-specific risk factors, such as breast density and background parenchymal enhancement (BPE), risk-stratified screening can help identify high-risk women and provide personalized screening plans, ultimately leading to better outcomes. Measurements of density and BPE are carried out through image segmentation, but volumetric measurements may not capture the qualitative scale of these tissue-specific risk factors. This study aimed to create deep regression models that estimate the interval scale underlying the BI-RADS density and BPE categories. These models incorporate a 3D convolutional encoder and transformer layers to comprehend time-sequential data in DCE-MRI. The correlation between the models and the BI-RADS categories was evaluated with Spearman coefficients. Using 1024 patients with a BI-RADS assessment score of 3 or less and no prior history of breast cancer, the models were trained on 50% of the data and tested on 50%. The density and BPE ground truth labels were extracted from the radiology reports using BI-RADS BERT. The ordinal classes were then translated to a continuous interval scale using a linear link function. The density regression model is strongly correlated to the BI-RADS category with a correlation of 0.77, slightly lower than segmentation %FGT. The BPE regression model with transformer layers shows a moderate correlation with radiologists at 0.52, similar to the segmentation %BPE. The deep regression transformer has an advantage over segmentation as it doesn’t need time-point image registration, making it easier to use.
(2024) Published by SPIE. Downloading of the abstract is permitted for personal use only.
Citation Download Citation
Grey Kuling, Belinda Curpen, and Anne L . Martel "Accurate estimation of density and background parenchymal enhancement in breast MRI using deep regression and transformers" |
Hesse, Linde S; Kuling, Grey; Veta, Mitko; Martel, Anne L.: Intensity Augmentation to Improve Generalizability of Breast Segmentation Across Different MRI Scan Protocols. In: IEEE Transactions on Biomedical Engineering, vol. 68, no. 3, pp. 759–770, 2021, ISSN: 0018-9294. @article{Hesse2020,
title = {Intensity Augmentation to Improve Generalizability of Breast Segmentation Across Different MRI Scan Protocols},
author = {Linde S Hesse and Grey Kuling and Mitko Veta and Anne L. Martel},
url = {https://ieeexplore.ieee.org/document/9166708/},
doi = {10.1109/TBME.2020.3016602},
issn = {0018-9294},
year = {2021},
date = {2021-03-01},
urldate = {2021-03-01},
journal = {IEEE Transactions on Biomedical Engineering},
volume = {68},
number = {3},
pages = {759–770},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Kuling, Grey; Curpen, Belinda; Martel, Anne L.: Domain adapted breast tissue segmentation in magnetic resonance imaging. In: Ongeval, Chantal Van; Marshall, Nicholas; Bosmans, Hilde (Ed.): 15th International Workshop on Breast Imaging (IWBI2020), pp. 61, SPIE, 2020, ISBN: 9781510638310. @inproceedings{Kuling2020,
title = {Domain adapted breast tissue segmentation in magnetic resonance imaging},
author = {Grey Kuling and Belinda Curpen and Anne L. Martel},
editor = {Chantal Van Ongeval and Nicholas Marshall and Hilde Bosmans},
url = {https://doi.org/10.1117/12.2564131 https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11513/2564131/Domain-adapted-breast-tissue-segmentation-in-magnetic-resonance-imaging/10.1117/12.2564131.full},
doi = {10.1117/12.2564131},
isbn = {9781510638310},
year = {2020},
date = {2020-05-01},
urldate = {2020-05-01},
booktitle = {15th International Workshop on Breast Imaging (IWBI2020)},
pages = {61},
publisher = {SPIE},
abstract = {For women of high risk ($>25%$ lifetime risk) for developing Breast Cancer combination screening of mammography and magnetic resonance imaging (MRI) is recommended. Risk stratification is based on current modeling tools for risk assessment. However, adding additional radiological features may improve AUC. To validate tissue features in MRI requires large scale epidemiological studies across health centres. Therefore it is essential to have a robust, fully automated segmentation method. This presents a challenge of imaging domain adaptation in deep learning. Here, we present a breast segmentation pipeline that uses multiple UNet segmentation models trained on different image types. We use Monte-Carlo Dropout to measure each model's uncertainty allowing the most appropriate model to be selected when the image domain is unknown. We show our pipeline achieves a dice similarity average of 0.78 for fibroglandular tissue segmentation and has good adherence to radiologist assessment.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
For women of high risk ($>25%$ lifetime risk) for developing Breast Cancer combination screening of mammography and magnetic resonance imaging (MRI) is recommended. Risk stratification is based on current modeling tools for risk assessment. However, adding additional radiological features may improve AUC. To validate tissue features in MRI requires large scale epidemiological studies across health centres. Therefore it is essential to have a robust, fully automated segmentation method. This presents a challenge of imaging domain adaptation in deep learning. Here, we present a breast segmentation pipeline that uses multiple UNet segmentation models trained on different image types. We use Monte-Carlo Dropout to measure each model's uncertainty allowing the most appropriate model to be selected when the image domain is unknown. We show our pipeline achieves a dice similarity average of 0.78 for fibroglandular tissue segmentation and has good adherence to radiologist assessment. |
Fashandi, Homa; Kuling, Grey; Lu, YingLi; Wu, Hongbo; Martel, Anne L.: An investigation of the effect of fat suppression and dimensionality on the accuracy of breast MRI segmentation using U-nets. In: Medical Physics, 2019, (This is the pre-peer reviewed version. The definitive version is available at: https://aapm.onlinelibrary.wiley.com/doi/abs/10.1002/mp.13375). @article{Fashandi2019,
title = {An investigation of the effect of fat suppression and dimensionality on the accuracy of breast MRI segmentation using U-nets},
author = {Homa Fashandi and Grey Kuling and YingLi Lu and Hongbo Wu and Anne L. Martel},
url = {http://hdl.handle.net/1807/93313},
doi = {10.1002/mp.13375},
year = {2019},
date = {2019-01-04},
urldate = {2019-01-04},
journal = {Medical Physics},
abstract = {Purpose
Accurate segmentation of the breast is required for breast density estimation and the assessment of background parenchymal enhancement, both of which have been shown to be related to breast cancer risk. The MRI breast segmentation task is challenging, and recent work has demonstrated that convolutional neural networks perform well for this task. In this study, we have investigated the performance of several 2D U‐Net and 3D U‐Net configurations using both fat‐suppressed and nonfat suppressed images. We have also assessed the effect of changing the number and quality of the ground truth segmentations.
Materials and methods
We designed 8 studies to investigate the effect of input types and the dimensionality of the U‐Net operations for the breast MRI segmentation. Our training data contained 70 whole breast volumes of T1‐weighted sequences without fat suppression(WOFS) and with fat suppression(FS). For each subject, we registered the WOFS and FS volumes together before manually segmenting the breast to generate ground truth. We compared 4 different input types to the U‐nets: WOFS, FS, MIXED(WOFS and FS images treated as separate samples) and MULTI(WOFS and FS images combined into a single multi‐channel image). We trained 2D U‐Nets and 3D U‐Nets with this data, which resulted in our 8 studies (2D‐WOFS, 3D‐WOFS,2D‐FS,3D‐FS,2D‐MIXED,3D‐MIXED,2D‐MULTI, and 3D‐MULT). For each of these studies, we performed a systematic grid search to tune the hyperparameters of the U‐Nets. A separate validation set with 15 whole breast volumes was used for hyperparameter tuning. We performed Kruskal‐Walis test on the results of our hyperparameter tuning and did not find a statistically significant difference in the 10 top models of each study. For this reason, we chose the best model as the model with the highest mean Dice Similarity Coefficient(DSC) value on the validation set. The reported test results are the results of the top model of each study on our test set which contained 19 whole breast volumes annotated by 3 readers fused with the STAPLE algorithm. We also investigated the effect of the quality of the training annotations and the number of training samples for this task.
Results
The study with the highest average DSC result was 3D‐MULTI with 0.96 ± 0.02. The second highest average is 2D WOFS (0.96 ± 0.03), and the third is 2D MULTI (0.96 ± 0.03). We performed the Kruskal‐Wallis 1‐way ANOVA test with Dunn's multiple comparison tests using Bonferroni p‐value correction on the results of the selected model of each study and found that 3D‐MULTI, 2D‐MULTI, 3D‐WOFS, 2D‐WOFS, 2D‐FS, and 3D‐FS were not statistically different in their distributions, which indicates that comparable results could be obtained in fat‐suppressed and nonfat suppressed volumes and that there is no significant difference between the 3D and 2D approach. Our results also suggested that the networks trained on single sequence images or multiple sequence images organized in multi‐channel images perform better than the models trained on a mixture of volumes from different sequences. Our investigation of the size of the training set revealed that training a U‐Net in this domain only requires a modest amount of training data and results obtained with 49 and 70 training datasets were not significantly different.
Conclusions
To summarize, we investigated the use of 2D U‐Nets and 3D U‐Nets for breast volume segmentation in T1 fat suppressed and without fat suppressed volumes. Although our highest score was obtained in the 3D MULTI study, when we took advantage of information in both fat suppressed and non fat suppressed volumes and their 3D structure, all of the methods we explored gave accurate segmentations with an average DSC on > 94% demonstrating that the U‐Net is a robust segmentation method for breast MRI volumes.},
note = {This is the pre-peer reviewed version. The definitive version is available at: https://aapm.onlinelibrary.wiley.com/doi/abs/10.1002/mp.13375},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Purpose
Accurate segmentation of the breast is required for breast density estimation and the assessment of background parenchymal enhancement, both of which have been shown to be related to breast cancer risk. The MRI breast segmentation task is challenging, and recent work has demonstrated that convolutional neural networks perform well for this task. In this study, we have investigated the performance of several 2D U‐Net and 3D U‐Net configurations using both fat‐suppressed and nonfat suppressed images. We have also assessed the effect of changing the number and quality of the ground truth segmentations.
Materials and methods
We designed 8 studies to investigate the effect of input types and the dimensionality of the U‐Net operations for the breast MRI segmentation. Our training data contained 70 whole breast volumes of T1‐weighted sequences without fat suppression(WOFS) and with fat suppression(FS). For each subject, we registered the WOFS and FS volumes together before manually segmenting the breast to generate ground truth. We compared 4 different input types to the U‐nets: WOFS, FS, MIXED(WOFS and FS images treated as separate samples) and MULTI(WOFS and FS images combined into a single multi‐channel image). We trained 2D U‐Nets and 3D U‐Nets with this data, which resulted in our 8 studies (2D‐WOFS, 3D‐WOFS,2D‐FS,3D‐FS,2D‐MIXED,3D‐MIXED,2D‐MULTI, and 3D‐MULT). For each of these studies, we performed a systematic grid search to tune the hyperparameters of the U‐Nets. A separate validation set with 15 whole breast volumes was used for hyperparameter tuning. We performed Kruskal‐Walis test on the results of our hyperparameter tuning and did not find a statistically significant difference in the 10 top models of each study. For this reason, we chose the best model as the model with the highest mean Dice Similarity Coefficient(DSC) value on the validation set. The reported test results are the results of the top model of each study on our test set which contained 19 whole breast volumes annotated by 3 readers fused with the STAPLE algorithm. We also investigated the effect of the quality of the training annotations and the number of training samples for this task.
Results
The study with the highest average DSC result was 3D‐MULTI with 0.96 ± 0.02. The second highest average is 2D WOFS (0.96 ± 0.03), and the third is 2D MULTI (0.96 ± 0.03). We performed the Kruskal‐Wallis 1‐way ANOVA test with Dunn's multiple comparison tests using Bonferroni p‐value correction on the results of the selected model of each study and found that 3D‐MULTI, 2D‐MULTI, 3D‐WOFS, 2D‐WOFS, 2D‐FS, and 3D‐FS were not statistically different in their distributions, which indicates that comparable results could be obtained in fat‐suppressed and nonfat suppressed volumes and that there is no significant difference between the 3D and 2D approach. Our results also suggested that the networks trained on single sequence images or multiple sequence images organized in multi‐channel images perform better than the models trained on a mixture of volumes from different sequences. Our investigation of the size of the training set revealed that training a U‐Net in this domain only requires a modest amount of training data and results obtained with 49 and 70 training datasets were not significantly different.
Conclusions
To summarize, we investigated the use of 2D U‐Nets and 3D U‐Nets for breast volume segmentation in T1 fat suppressed and without fat suppressed volumes. Although our highest score was obtained in the 3D MULTI study, when we took advantage of information in both fat suppressed and non fat suppressed volumes and their 3D structure, all of the methods we explored gave accurate segmentations with an average DSC on > 94% demonstrating that the U‐Net is a robust segmentation method for breast MRI volumes. |
Kuling, Grey; Fashandi, Homa; Lu, YingLi; Wu, Hongbo; Martel, Anne L.: Breast Volume and Fibroglandular Tissue Segmentation in MRI using a Deep Learning Unet. ISMRM Workshop on Breast MRI: Advancing the State of the Art, 2018. @workshop{Kuling2018,
title = {Breast Volume and Fibroglandular Tissue Segmentation in MRI using a Deep Learning Unet},
author = {Grey Kuling and Homa Fashandi and YingLi Lu and Hongbo Wu and Anne L. Martel},
url = {http://martellab.com/wp-content/uploads/2019/09/GCK_ISMRMAbstract_DLSegmatation_072018-3.pdf},
year = {2018},
date = {2018-09-10},
urldate = {2018-09-10},
booktitle = {ISMRM Workshop on Breast MRI: Advancing the State of the Art},
keywords = {},
pubstate = {published},
tppubtype = {workshop}
}
|